`datasummary`: Crosstabs, frequencies, correlations, balance (a.k.a. "table 1"), and more
Source:vignettes/datasummary.Rmd
datasummary.Rmddatasummary is a function from the
modelsummary package. It allows us to create data
summaries, frequency tables, crosstabs, correlation tables, balance
tables (aka “Table 1”), and more. It has many benefits:
- Easy to use.
- Extremely flexible.
- Many output formats: HTML, LaTeX, Microsoft Word and Powerpoint, Text/Markdown, PDF, RTF, or Image files.
- Embed tables in
Rmarkdownorknitrdynamic documents. -
Customize the appearance of tables with the
gt,kableExtraorflextablepackages. The possibilities are endless!
This tutorial will show how to draw tables like these (and more!):
Background
datasummary is built around the fantastic tables
package for R. It is a thin “wrapper” which adds
convenience functions and arguments; a user-interface consistent with
modelsummary; cleaner html output; and the ability to
export tables to more formats, including gt tables, flextable
objects, and Microsoft Word documents.
datasummary is a general-purpose table-making tool. It
allows us to build (nearly) any summary table we want by using simple 2-sided formulae. For example, in the
expression x + y ~ mean + sd, the left-hand side of the
formula identifies the variables or statistics to display as rows, and
the right-hand side defines the columns. Below, we will see how
variables and statistics can be “nested” with the *
operator to produce tables like the ones above.
In addition to datasummary, the
modelsummary package includes a “family” of companion
functions named datasummary_*. These functions facilitate
the production of standard, commonly used tables. This family currently
includes:
-
datasummary(): Flexible function to create custom tables using 2-sided formulae. -
datasummary_balance(): Group characteristics (e.g., control vs. treatment) -
datasummary_correlation(): Table of correlations. -
datasummary_skim(): Quick summary of a dataset. -
datasummary_df(): Create a table from any dataframe. -
datasummary_crosstab(): Cross tabulations of categorical variables.
In the next three sections, we illustrate how to use
datasummary_balance, datasummary_correlation,
datasummary_skim, and datasummary_crosstab.
Then, we dive into datasummary itself to highlight its ease
and flexibility.
datasummary_skim
The first datasummary companion function is called
datasummary_skim. It was heavily inspired by one
of my favorite data exploration tools for R: the skimr package.
The goal of this function is to give us a quick look at the data.
To illustrate, we download data from the cool new palmerpenguins
package by Alison Presmanes Hill and Allison Horst. These data were
collected at the Palmer Station in Antarctica by Gorman, Williams &
Fraser (2014), and they include 3 categorical variables and 4 numeric
variables.
library(modelsummary)
library(tidyverse)
url <- 'https://vincentarelbundock.github.io/Rdatasets/csv/palmerpenguins/penguins.csv'
penguins <- read.csv(url, na.strings = "")To summarize the numeric variables in the dataset, we type:
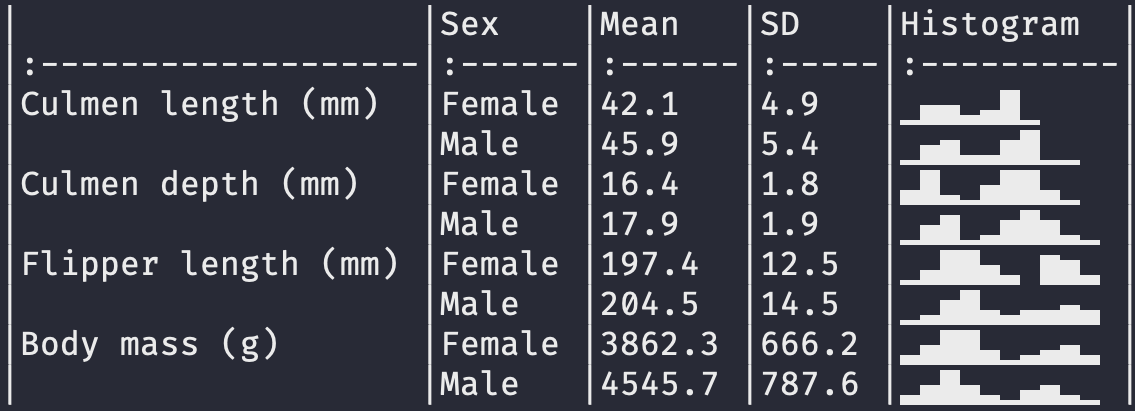
datasummary_skim(penguins)
To summarize the categorical variables in the dataset, we type:
datasummary_skim(penguins, type = "categorical")| N | % | ||
|---|---|---|---|
| species | Adelie | 152 | 44.2 |
| Chinstrap | 68 | 19.8 | |
| Gentoo | 124 | 36.0 | |
| island | Biscoe | 168 | 48.8 |
| Dream | 124 | 36.0 | |
| Torgersen | 52 | 15.1 | |
| sex | female | 165 | 48.0 |
| male | 168 | 48.8 | |
| NA | 11 | 3.2 |
Later in this tutorial, it will become clear that
datasummary_skim is just a convenience “template” built
around datasummary, since we can achieve identical results
with the latter. For example, to produce a text-only version of the
tables above, we can type:
datasummary(All(penguins) ~ Mean + SD + Histogram,
data = penguins,
output = 'markdown')| | Mean| SD| Histogram|
|:-----------------|-------:|------:|----------:|
|bill_length_mm | 43.92| 5.46| ▁▅▆▆▆▇▇▂▁|
|bill_depth_mm | 17.15| 1.97| ▃▄▄▄▇▆▇▅▂▁|
|flipper_length_mm | 200.92| 14.06| ▂▅▇▄▁▄▄▂▁|
|body_mass_g | 4201.75| 801.95| ▁▄▇▅▄▄▃▃▂▁|Printing histograms will not work on all computers. If you have issues with this feature, try changing your computer’s locale, or try using a different display font.
The datasummary_skim function does not currently allow
users to summarize continuous and categorical variables together in a
single table, but the datasummary_balance function
described in the next section can do so.
datasummary_balance
The expressions “balance table” or “Table 1” refer to a type of table which is often printed in the opening pages of a scientific peer-reviewed article. Typically, this table includes basic descriptive statistics about different subsets of the study population. For instance, analysts may want to compare the socio-demographic characteristics of members of the “control” and “treatment” groups in a randomized control trial, or the flipper lengths of male and female penguins. In addition, balance tables often include difference in means tests.
To illustrate how to build a balance table using the
datasummary_balance function, we download data about a job
training experiment studies in Lalonde (1986). Then, we clean up the
data by renaming and recoding a few variables.
# Download and read data
training <- 'https://vincentarelbundock.github.io/Rdatasets/csv/Ecdat/Treatment.csv'
training <- read.csv(training, na.strings = "")
# Rename and recode variables
training <- training %>%
mutate(`Earnings Before` = re75 / 1000,
`Earnings After` = re78 / 1000,
Treatment = ifelse(treat == TRUE, 'Treatment', 'Control'),
Married = ifelse(married == TRUE, 'Yes', 'No')) %>%
select(`Earnings Before`,
`Earnings After`,
Treatment,
Ethnicity = ethn,
Age = age,
Education = educ,
Married)Now, we execute the datasummary_balance function. If the
estimatr package is installed,
datasummary_balance will calculate the difference in means
and test statistics.
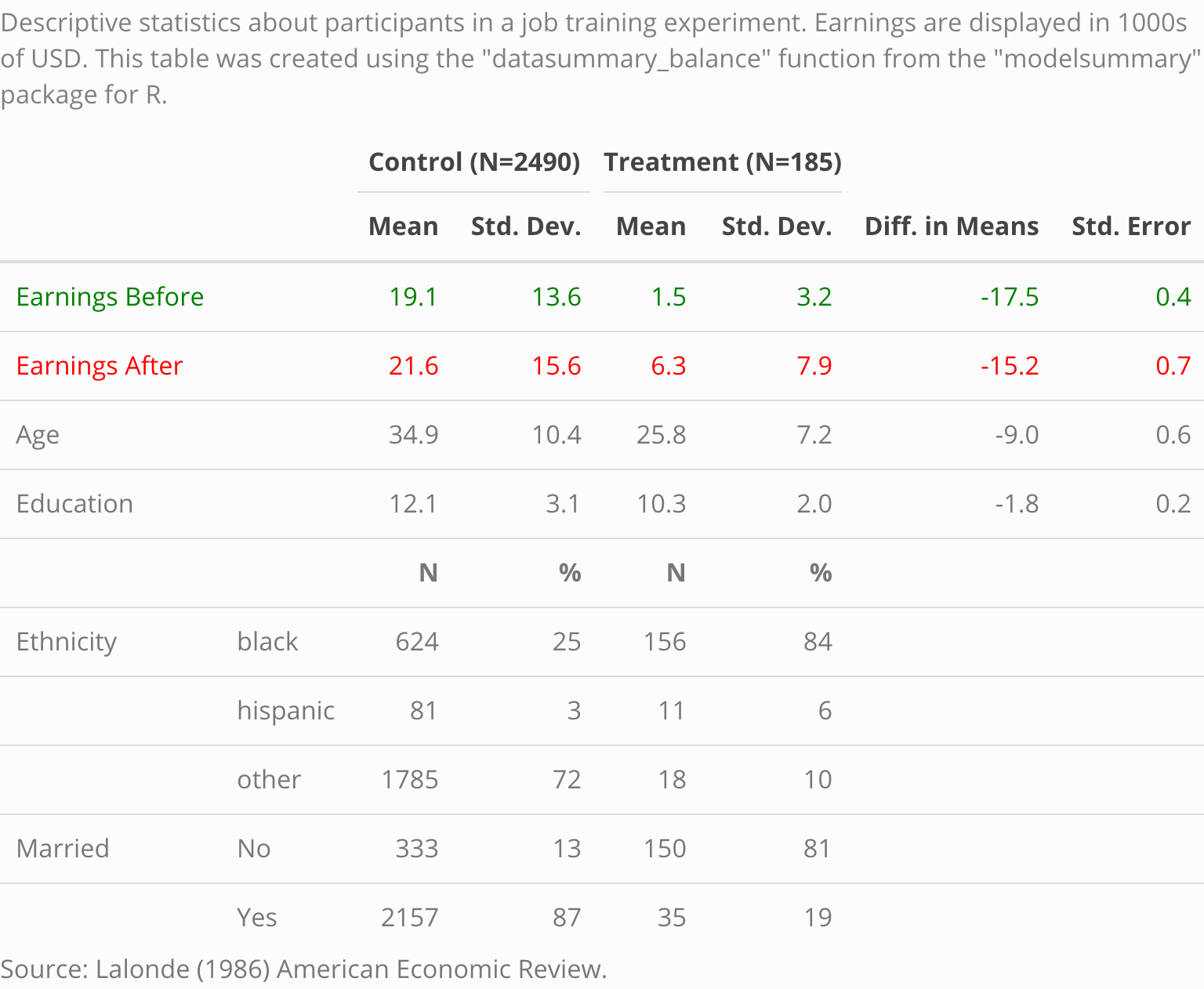
caption <- 'Descriptive statistics about participants in a job training experiment. The earnings are displayed in 1000s of USD. This table was created using the "datasummary" function from the "modelsummary" package for R.'
reference <- 'Source: Lalonde (1986) American Economic Review.'
library(modelsummary)
datasummary_balance(~Treatment,
data = training,
title = caption,
notes = reference)Note that if the dataset includes columns called “blocks”,
“clusters”, or “weights”, this information will automatically be taken
into consideration by estimatr when calculating the
difference in means and the associated statistics.
Users can also use the ~ 1 formula to indicate that they
want to summarize all the data instead of splitting the analysis across
subgroups:
datasummary_balance(~ 1, data = training)| Mean | Std. Dev. | ||
|---|---|---|---|
| Earnings Before | 17.9 | 13.9 | |
| Earnings After | 20.5 | 15.6 | |
| Age | 34.2 | 10.5 | |
| Education | 12.0 | 3.1 | |
| N | Pct. | ||
| Treatment | Control | 2490 | 93.1 |
| Treatment | 185 | 6.9 | |
| Ethnicity | black | 780 | 29.2 |
| hispanic | 92 | 3.4 | |
| other | 1803 | 67.4 | |
| Married | No | 483 | 18.1 |
| Yes | 2192 | 81.9 |
datasummary_correlation
The datasummary_correlation accepts a dataframe or
tibble, it identifies all the numeric variables, and calculates the
correlation between each of those variables:
datasummary_correlation(mtcars)| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| mpg | 1 | . | . | . | . | . | . | . | . | . | . |
| cyl | -.85 | 1 | . | . | . | . | . | . | . | . | . |
| disp | -.85 | .90 | 1 | . | . | . | . | . | . | . | . |
| hp | -.78 | .83 | .79 | 1 | . | . | . | . | . | . | . |
| drat | .68 | -.70 | -.71 | -.45 | 1 | . | . | . | . | . | . |
| wt | -.87 | .78 | .89 | .66 | -.71 | 1 | . | . | . | . | . |
| qsec | .42 | -.59 | -.43 | -.71 | .09 | -.17 | 1 | . | . | . | . |
| vs | .66 | -.81 | -.71 | -.72 | .44 | -.55 | .74 | 1 | . | . | . |
| am | .60 | -.52 | -.59 | -.24 | .71 | -.69 | -.23 | .17 | 1 | . | . |
| gear | .48 | -.49 | -.56 | -.13 | .70 | -.58 | -.21 | .21 | .79 | 1 | . |
| carb | -.55 | .53 | .39 | .75 | -.09 | .43 | -.66 | -.57 | .06 | .27 | 1 |
The values displayed in this table are equivalent to those obtained
by calling: cor(x, use='pairwise.complete.obs').
datasummary_crosstab
A cross tabulation is often useful to explore the association between two categorical variables.
library(modelsummary)
url <- 'https://vincentarelbundock.github.io/Rdatasets/csv/palmerpenguins/penguins.csv'
penguins <- read.csv(url, na.strings = "")
datasummary_crosstab(species ~ sex, data = penguins)| species | female | male | All | |
|---|---|---|---|---|
| Adelie | N | 73 | 73 | 152 |
| % row | 48.0 | 48.0 | 100.0 | |
| Chinstrap | N | 34 | 34 | 68 |
| % row | 50.0 | 50.0 | 100.0 | |
| Gentoo | N | 58 | 61 | 124 |
| % row | 46.8 | 49.2 | 100.0 | |
| All | N | 165 | 168 | 344 |
| % row | 48.0 | 48.8 | 100.0 |
You can create multi-level crosstabs by specifying interactions using
the * operator:
datasummary_crosstab(species ~ sex * island, data = penguins)| species | female | male | All | |||||
|---|---|---|---|---|---|---|---|---|
| Biscoe | Dream | Torgersen | Biscoe | Dream | Torgersen | |||
| Adelie | N | 22 | 27 | 24 | 22 | 28 | 23 | 152 |
| % row | 14.5 | 17.8 | 15.8 | 14.5 | 18.4 | 15.1 | 100.0 | |
| Chinstrap | N | 0 | 34 | 0 | 0 | 34 | 0 | 68 |
| % row | 0.0 | 50.0 | 0.0 | 0.0 | 50.0 | 0.0 | 100.0 | |
| Gentoo | N | 58 | 0 | 0 | 61 | 0 | 0 | 124 |
| % row | 46.8 | 0.0 | 0.0 | 49.2 | 0.0 | 0.0 | 100.0 | |
| All | N | 80 | 61 | 24 | 83 | 62 | 23 | 344 |
| % row | 23.3 | 17.7 | 7.0 | 24.1 | 18.0 | 6.7 | 100.0 | |
By default, the cell counts and row percentages are shown for each
cell, and both row and column totals are calculated. To show cell
percentages or column percentages, or to drop row and column totals,
adjust the statistic argument. This argument accepts a
formula that follows the datasummary “language”. To
understand exactly how it works, you may find it useful to skip to the
datasummary tutorial in the next section. Example:
datasummary_crosstab(species ~ sex,
statistic = 1 ~ Percent("col"),
data = penguins)| species | female | male | |
|---|---|---|---|
| Adelie | % col | 44.2 | 43.5 |
| Chinstrap | % col | 20.6 | 20.2 |
| Gentoo | % col | 35.2 | 36.3 |
| All | % col | 100.0 | 100.0 |
See ?datasummary_crosstab for more details.
datasummary
datasummary tables are specified using a 2-sided
formula, divided by a tilde ~. The left-hand side describes
the rows; the right-hand side describes the columns. To illustrate how
this works, we will again be using the palmerpenguins dataset:
To display the flipper_length_mm variable as a row and
the mean as a column, we type:
datasummary(flipper_length_mm ~ Mean,
data = penguins)| Mean | |
|---|---|
| flipper_length_mm | 200.92 |
To flip rows and columns, we flip the left and right-hand sides of the formula:
datasummary(Mean ~ flipper_length_mm,
data = penguins)| flipper_length_mm | |
|---|---|
| Mean | 200.92 |
Custom summary functions
The Mean function is a shortcut supplied by
modelsummary, and it is equivalent to
mean(x,na.rm=TRUE). Since the
flipper_length_mm variable includes missing observation,
using the mean formula (with default
na.rm=FALSE) would produce a missing/empty cell:
datasummary(flipper_length_mm ~ mean,
data = penguins)| mean | |
|---|---|
| flipper_length_mm |
modelsummary supplies these functions:
Mean, SD, Min, Max,
Median, P0, P25,
P50, P75, P100,
Histogram, and a few more (see the package
documentation).
Users are also free to create and use their own custom summaries. Any
R function which takes a vector and produces a single value
is acceptable. For example, the Range functions return a
numerical value, and the MinMax returns a string:
Range <- function(x) max(x, na.rm = TRUE) - min(x, na.rm = TRUE)
datasummary(flipper_length_mm ~ Range,
data = penguins)| Range | |
|---|---|
| flipper_length_mm | 59.00 |
MinMax <- function(x) paste0('[', min(x, na.rm = TRUE), ', ', max(x, na.rm = TRUE), ']')
datasummary(flipper_length_mm ~ MinMax,
data = penguins)| MinMax | |
|---|---|
| flipper_length_mm | [172, 231] |
Concatenating with +
To include more rows and columns, we use the + sign:
datasummary(flipper_length_mm + body_mass_g ~ Mean + SD,
data = penguins)| Mean | SD | |
|---|---|---|
| flipper_length_mm | 200.92 | 14.06 |
| body_mass_g | 4201.75 | 801.95 |
Sometimes, it can be cumbersome to list all variables separated by
+ signs. The All() function is a useful
shortcut:
datasummary(All(penguins) ~ Mean + SD,
data = penguins)| Mean | SD | |
|---|---|---|
| rownames | 172.50 | 99.45 |
| bill_length_mm | 43.92 | 5.46 |
| bill_depth_mm | 17.15 | 1.97 |
| flipper_length_mm | 200.92 | 14.06 |
| body_mass_g | 4201.75 | 801.95 |
| year | 2008.03 | 0.82 |
By default, All selects all numeric variables. This
behavior can be changed by modifying the function’s arguments. See
?All for details.
Nesting with *
datasummary can nest variables and statistics inside
categorical variables using the * symbol. When applying the
the * operator to factor, character, or logical variables,
columns or rows will automatically be nested. For instance, if we want
to display separate means for each value of the variable
sex, we use mean * sex:
datasummary(flipper_length_mm + body_mass_g ~ mean * sex,
data = penguins)| female | male | |
|---|---|---|
| flipper_length_mm | 197.36 | 204.51 |
| body_mass_g | 3862.27 | 4545.68 |
We can use parentheses to nest several terms inside one another,
using a call of this form: x * (y + z). Here is an example
with nested columns:
datasummary(body_mass_g ~ sex * (mean + sd),
data = penguins)| female | male | |||
|---|---|---|---|---|
| mean | sd | mean | sd | |
| body_mass_g | 3862.27 | 666.17 | 4545.68 | 787.63 |
Here is an example with nested rows:
datasummary(sex * (body_mass_g + flipper_length_mm) ~ mean + sd,
data = penguins)| sex | mean | sd | |
|---|---|---|---|
| female | body_mass_g | 3862.27 | 666.17 |
| flipper_length_mm | 197.36 | 12.50 | |
| male | body_mass_g | 4545.68 | 787.63 |
| flipper_length_mm | 204.51 | 14.55 |
The order in which terms enter the formula determines the order in
which labels are displayed. For example, this shows island
above sex:
datasummary(flipper_length_mm + body_mass_g ~ mean * island * sex,
data = penguins)| Biscoe | Dream | Torgersen | ||||
|---|---|---|---|---|---|---|
| female | male | female | male | female | male | |
| flipper_length_mm | 205.69 | 213.29 | 190.02 | 196.31 | 188.29 | 194.91 |
| body_mass_g | 4319.38 | 5104.52 | 3446.31 | 3987.10 | 3395.83 | 4034.78 |
This shows sex above island values:
datasummary(flipper_length_mm + body_mass_g ~ mean * sex * island,
data = penguins)| female | male | |||||
|---|---|---|---|---|---|---|
| Biscoe | Dream | Torgersen | Biscoe | Dream | Torgersen | |
| flipper_length_mm | 205.69 | 190.02 | 188.29 | 213.29 | 196.31 | 194.91 |
| body_mass_g | 4319.38 | 3446.31 | 3395.83 | 5104.52 | 3987.10 | 4034.78 |
By default, datasummary omits column headers with a
single value/label across all columns, except for the header that sits
just above the data. If the header we want to see is not displayed, we
may want to reorder the terms of the formula. To show all headers, set
sparse_header=FALSE:
datasummary(flipper_length_mm + body_mass_g ~ mean * sex * island,
data = penguins,
sparse_header = FALSE)| female | male | |||||
|---|---|---|---|---|---|---|
| Biscoe | Dream | Torgersen | Biscoe | Dream | Torgersen | |
| flipper_length_mm | 205.69 | 190.02 | 188.29 | 213.29 | 196.31 | 194.91 |
| body_mass_g | 4319.38 | 3446.31 | 3395.83 | 5104.52 | 3987.10 | 4034.78 |
When using sparse_header=FALSE, it is often useful to
insert Heading() * in the table formula, in order to rename
or omit some of the labels manually. Type ?tables::Heading
for details and examples.
Renaming with =
Personally, I prefer to rename variables and values before drawing my tables, using backticks when variable names include whitespace. For example,
tmp <- penguins %>%
select(`Flipper length (mm)` = flipper_length_mm,
`Body mass (g)` = body_mass_g)
datasummary(`Flipper length (mm)` + `Body mass (g)` ~ Mean + SD,
data = tmp)| Mean | SD | |
|---|---|---|
| Flipper length (mm) | 200.92 | 14.06 |
| Body mass (g) | 4201.75 | 801.95 |
However, thanks to the tables package,
datasummary offers two additional mechanisms to rename.
First, we can wrap a term in parentheses and use the equal
= sign: (NewName=OldName):
datasummary((`Flipper length (mm)` = flipper_length_mm) + (`Body mass (g)` = body_mass_g) ~
island * ((Avg. = Mean) + (Std.Dev. = SD)),
data = penguins)| Biscoe | Dream | Torgersen | ||||
|---|---|---|---|---|---|---|
| Avg. | Std.Dev. | Avg. | Std.Dev. | Avg. | Std.Dev. | |
| Flipper length (mm) | 209.71 | 14.14 | 193.07 | 7.51 | 191.20 | 6.23 |
| Body mass (g) | 4716.02 | 782.86 | 3712.90 | 416.64 | 3706.37 | 445.11 |
Second, we can use the Heading() function:
datasummary(Heading("Flipper length (mm)") * flipper_length_mm + Heading("Body mass (g)") * body_mass_g ~ island * (Mean + SD),
data = penguins)| Biscoe | Dream | Torgersen | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Flipper length (mm) | 209.71 | 14.14 | 193.07 | 7.51 | 191.20 | 6.23 |
| Body mass (g) | 4716.02 | 782.86 | 3712.90 | 416.64 | 3706.37 | 445.11 |
The Heading function also has a nearData
argument which can be useful in cases where some rows are nested but
others are not. Compare the last row of these two tables:
datasummary(sex * (flipper_length_mm + bill_length_mm) + Heading("Body mass (g)") * body_mass_g ~ Mean + SD,
data = penguins)| sex | Mean | SD | |
|---|---|---|---|
| female | flipper_length_mm | 197.36 | 12.50 |
| bill_length_mm | 42.10 | 4.90 | |
| male | flipper_length_mm | 204.51 | 14.55 |
| bill_length_mm | 45.85 | 5.37 | |
| Body mass (g) | 4201.75 | 801.95 |
datasummary(sex * (flipper_length_mm + bill_length_mm) + Heading("Body mass (g)", nearData=FALSE) * body_mass_g ~ Mean + SD,
data = penguins)| sex | Mean | SD | |
|---|---|---|---|
| female | flipper_length_mm | 197.36 | 12.50 |
| bill_length_mm | 42.10 | 4.90 | |
| male | flipper_length_mm | 204.51 | 14.55 |
| bill_length_mm | 45.85 | 5.37 | |
| Body mass (g) | 4201.75 | 801.95 |
Counts and Percentages
The tables package allows datasummary to
use neat tricks to produce frequency tables:
- Add a
Nto the right-hand side of the equation. - Add
Percent()to the right-hand side to calculate the percentage of observations in each cell. - Add
1to the left-hand side to include a row with the total number of observations:
datasummary(species * sex + 1 ~ N + Percent(),
data = penguins)| species | sex | N | Percent |
|---|---|---|---|
| Adelie | female | 73 | 21.22 |
| male | 73 | 21.22 | |
| Chinstrap | female | 34 | 9.88 |
| male | 34 | 9.88 | |
| Gentoo | female | 58 | 16.86 |
| male | 61 | 17.73 | |
| All | 344 | 100.00 |
Note that the Percent() function accepts a
denom argument to determine if percentages should be
calculated row or column-wise, or if they should take into account all
cells.
Weighted percentages
The Percent() pseudo-function also accepts a
fn argument, which must be a function which accepts two
vectors: x is the values in the current cell, and
y is all the values in the whole dataset. The default
fn is:
datasummary(species * sex + 1 ~ N + Percent(fn = function(x, y) 100 * length(x) / length(y)),
output = "markdown",
data = penguins)| species | sex | N | Percent |
|---|---|---|---|
| Adelie | female | 73 | 21.22 |
| male | 73 | 21.22 | |
| Chinstrap | female | 34 | 9.88 |
| male | 34 | 9.88 | |
| Gentoo | female | 58 | 16.86 |
| male | 61 | 17.73 | |
| All | 344 | 100.00 |
The code above takes the number of elements in the cell
length(x) and divides it by the number of total elements
length(y).
Now, let’s say we want to display percentages weighted by one of the
variables of the dataset. This can often be useful with survey weights,
for example. Here, we use an arbitrary column of weights called
flipper_length_mm:
wtpct <- function(x, y) sum(x, na.rm = TRUE) / sum(y, na.rm = TRUE) * 100
datasummary(species * sex + 1 ~ N + flipper_length_mm * Percent(fn = wtpct),
output = "markdown",
data = penguins)| species | sex | N | Percent |
|---|---|---|---|
| Adelie | female | 73 | 19.95 |
| male | 73 | 20.44 | |
| Chinstrap | female | 34 | 9.49 |
| male | 34 | 9.89 | |
| Gentoo | female | 58 | 17.95 |
| male | 61 | 19.67 | |
| All | 344 | 100.00 |
In each cell we now have the sum of weights in that cell, divided by the total sum of weights in the column.
Custom percentages
Here is another simple illustration of Percent function mechanism in action, where we combine counts and percentages in a simple nice label:
dat <- mtcars
dat$cyl <- as.factor(dat$cyl)
fn <- function(x, y) {
out <- sprintf(
"%s (%.1f%%)",
length(x),
length(x) / length(y) * 100)
}
datasummary(
cyl ~ Percent(fn = fn),
data = dat)| cyl | Percent |
|---|---|
| 4 | 11 (34.4%) |
| 6 | 7 (21.9%) |
| 8 | 14 (43.8%) |
Factor
The * nesting operator that we used above works
automatically for factor, character, and logical variables. Sometimes,
it is convenient to use it with other types of variables, such as binary
numeric variables. In that case, we can wrap the variable name inside a
call to Factor(). This allows us to treat a variable as
factor, without having to modify it in the original data. For example,
in the mtcars data, there is a binary numeric variable call
am. We nest statistics within categories of am
by typing:
datasummary(mpg + hp ~ Factor(am) * (mean + sd),
data = mtcars)| 0 | 1 | |||
|---|---|---|---|---|
| mean | sd | mean | sd | |
| mpg | 17.15 | 3.83 | 24.39 | 6.17 |
| hp | 160.26 | 53.91 | 126.85 | 84.06 |
Arguments: na.rm=TRUE
We can pass any argument to the summary function by including a call
to Arguments(). For instance, there are missing values in
the flipper_length_mm variable of the penguins
dataset. Therefore, the standard mean function will produce
no result, because its default argument is na.rm=FALSE. We
can change that by calling:
datasummary(flipper_length_mm ~ mean + mean*Arguments(na.rm=TRUE),
data = penguins)| mean | mean | |
|---|---|---|
| flipper_length_mm | 200.92 |
Notice that there is an empty cell (NA) under the normal
mean function, but that the mean call with
Arguments(na.rm=TRUE) produced a numeric result.
We can pass the same arguments to multiple functions using the parentheses:
datasummary(flipper_length_mm ~ (mean + sd) * Arguments(na.rm=TRUE),
data = penguins)| mean | sd | |
|---|---|---|
| flipper_length_mm | 200.92 | 14.06 |
Note that the shortcut functions that modelsummary
supplies use na.rm=TRUE by default, so we can use them
directly without arguments, even when there are missing values:
datasummary(flipper_length_mm ~ Mean + Var + P75 + Range,
data = penguins)| Mean | Var | P75 | Range | |
|---|---|---|---|---|
| flipper_length_mm | 200.92 | 197.73 | 213.00 | 59.00 |
Arguments: Weighted Mean
You can use the Arguments mechanism to do various
things, such as calculating weighted means:
newdata <- data.frame(
x = rnorm(20),
w = rnorm(20),
y = rnorm(20))
datasummary(x + y ~ weighted.mean * Arguments(w = w),
data = newdata, output = "markdown")| weighted.mean | |
|---|---|
| x | 0.70 |
| y | -0.47 |
Which produces the same results as:
weighted.mean(newdata$x, newdata$w)## [1] 0.6985158
weighted.mean(newdata$y, newdata$w)## [1] -0.4710503But different results from:
mean(newdata$x)## [1] -0.04014939
mean(newdata$y)## [1] -0.2950554Empty cells
Sometimes, if we nest too much and the dataset is not large/diverse
enough, we end up with empty cells. In that case, we add
*DropEmpty() to the formula:
datasummary(island * species * body_mass_g ~ sex * (Mean + SD),
data = penguins)| island | species | female | male | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Biscoe | Adelie | body_mass_g | 3369.32 | 343.47 | 4050.00 | 355.57 |
| Chinstrap | body_mass_g | |||||
| Gentoo | body_mass_g | 4679.74 | 281.58 | 5484.84 | 313.16 | |
| Dream | Adelie | body_mass_g | 3344.44 | 212.06 | 4045.54 | 330.55 |
| Chinstrap | body_mass_g | 3527.21 | 285.33 | 3938.97 | 362.14 | |
| Gentoo | body_mass_g | |||||
| Torgersen | Adelie | body_mass_g | 3395.83 | 259.14 | 4034.78 | 372.47 |
| Chinstrap | body_mass_g | |||||
| Gentoo | body_mass_g | |||||
datasummary(island * species * body_mass_g ~ sex * (Mean + SD) * DropEmpty(),
data = penguins)| island | species | female | male | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Biscoe | Adelie | body_mass_g | 3369.32 | 343.47 | 4050.00 | 355.57 |
| Gentoo | body_mass_g | 4679.74 | 281.58 | 5484.84 | 313.16 | |
| Dream | Adelie | body_mass_g | 3344.44 | 212.06 | 4045.54 | 330.55 |
| Chinstrap | body_mass_g | 3527.21 | 285.33 | 3938.97 | 362.14 | |
| Torgersen | Adelie | body_mass_g | 3395.83 | 259.14 | 4034.78 | 372.47 |
Logical subsets
Cool stuff is possible with logical subsets:
datasummary((bill_length_mm > 44.5) + (bill_length_mm <= 44.5) ~ Mean * flipper_length_mm,
data = penguins)| bill_length_mm > 44.5 | 209.68 |
| bill_length_mm <= 44.5 | 192.45 |
See the tables package documentation for more details
and examples.
output
All functions in the datasummary_* family accept the
same output argument. Tables can be saved to several file
formats:
f <- flipper_length_mm ~ island * (mean + sd)
datasummary(f, data = penguins, output = 'table.html')
datasummary(f, data = penguins, output = 'table.tex')
datasummary(f, data = penguins, output = 'table.docx')
datasummary(f, data = penguins, output = 'table.pptx')
datasummary(f, data = penguins, output = 'table.md')
datasummary(f, data = penguins, output = 'table.rtf')
datasummary(f, data = penguins, output = 'table.jpg')
datasummary(f, data = penguins, output = 'table.png')They can be returned in human-readable data.frames, markdown, html, or LaTeX code to the console:
datasummary(f, data = penguins, output = 'data.frame')
datasummary(f, data = penguins, output = 'markdown')
datasummary(f, data = penguins, output = 'html')
datasummary(f, data = penguins, output = 'latex')datasummary can return objects compatible with the
gt, kableExtra, flextable,
huxtable, and DT table creation and
customization packages:
datasummary(f, data = penguins, output = 'gt')
datasummary(f, data = penguins, output = 'kableExtra')
datasummary(f, data = penguins, output = 'flextable')
datasummary(f, data = penguins, output = 'huxtable')
datasummary(f, data = penguins, output = 'DT')Please note that hierarchical or “nested” column labels are only available for these output formats: kableExtra, gt, html, rtf, and LaTeX. When saving tables to other formats, nested labels will be combined to a “flat” header.
fmt
The fmt argument allows us to set the printing format of
numeric values. It accepts a single number representing the number of
digits after the period, or a string to be passed to the
sprintf function. For instance, setting
fmt="%.4f" will keep 4 digits after the dot (see
?sprintf for more options):
datasummary(flipper_length_mm ~ Mean + SD,
fmt = 4,
data = penguins)| Mean | SD | |
|---|---|---|
| flipper_length_mm | 200.9152 | 14.0617 |
We can set the formatting on a term-by-term basis by using the same
Arguments function that we used to handle missing values in
the previous section. The shortcut functions that ship with
modelsummary (e.g., Mean, SD,
Median, P25) all include a fmt
argument:
datasummary(flipper_length_mm ~ Mean * Arguments(fmt = "%.4f") + SD * Arguments(fmt = "%.1f"),
data = penguins)| Mean | SD | |
|---|---|---|
| flipper_length_mm | 200.9152 | 14.1 |
If we do not want datasummary to format
numbers, and we want to keep the numerical values instead of formatted
strings, set fmt=NULL. This can be useful when
post-processing tables with packages like gt, which allow
us to transform cells based on their numerical content (this
gt table will be omitted from PDF output):
library(gt)
datasummary(All(mtcars) ~ Mean + SD,
data = mtcars,
fmt = NULL,
output = 'gt') %>%
tab_style(style = cell_fill(color = "pink"),
locations = cells_body(rows = Mean > 10, columns = 2))| Mean | SD | |
|---|---|---|
| mpg | 20.090625 | 6.0269481 |
| cyl | 6.187500 | 1.7859216 |
| disp | 230.721875 | 123.9386938 |
| hp | 146.687500 | 68.5628685 |
| drat | 3.596563 | 0.5346787 |
| wt | 3.217250 | 0.9784574 |
| qsec | 17.848750 | 1.7869432 |
| vs | 0.437500 | 0.5040161 |
| am | 0.406250 | 0.4989909 |
| gear | 3.687500 | 0.7378041 |
| carb | 2.812500 | 1.6152000 |
Please note that the N() function is supplied by the
upstream tables package, and does not have a
fmt argument. Fortunately, it is easy to override the
built-in function to use custom formatting:
tmp <- data.frame(X = sample(letters[1:3], 1e6, replace = TRUE))
N <- \(x) format(length(x), big.mark = ",")
datasummary(X ~ N, data = tmp)| X | N |
|---|---|
| a | 332,358 |
| b | 334,065 |
| c | 333,577 |
title, notes
datasummary includes the same title and
notes arguments as in modelsummary:
datasummary(All(penguins) ~ Mean + SD,
data = penguins,
title = 'Statistics about the famous Palmer Penguins.',
notes = c('A note at the bottom of the table.'))| Mean | SD | |
|---|---|---|
| rownames | 172.50 | 99.45 |
| bill_length_mm | 43.92 | 5.46 |
| bill_depth_mm | 17.15 | 1.97 |
| flipper_length_mm | 200.92 | 14.06 |
| body_mass_g | 4201.75 | 801.95 |
| year | 2008.03 | 0.82 |
| A note at the bottom of the table. | ||
align
We can align columns using the align argument.
align should be a string of length equal to the number of
columns, and which includes only the letters “l”, “c”, or “r”:
datasummary(flipper_length_mm + bill_length_mm ~ Mean + SD + Range,
data = penguins,
align = 'lrcl')| Mean | SD | Range | |
|---|---|---|---|
| flipper_length_mm | 200.92 | 14.06 | 59.00 |
| bill_length_mm | 43.92 | 5.46 | 27.50 |
add_rows
new_rows <- data.frame('Does',
2,
'plus',
2,
'equals',
5,
'?')
datasummary(flipper_length_mm + body_mass_g ~ species * (Mean + SD),
data = penguins,
add_rows = new_rows)| Adelie | Chinstrap | Gentoo | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| flipper_length_mm | 189.95 | 6.54 | 195.82 | 7.13 | 217.19 | 6.48 |
| body_mass_g | 3700.66 | 458.57 | 3733.09 | 384.34 | 5076.02 | 504.12 |
| Does | 2.00 | plus | 2.00 | equals | 5.00 | ? |
add_columns
new_cols <- data.frame('New Stat' = runif(2))
datasummary(flipper_length_mm + body_mass_g ~ species * (Mean + SD),
data = penguins,
add_columns = new_cols)| Adelie | Chinstrap | Gentoo | New.Stat | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| flipper_length_mm | 189.95 | 6.54 | 195.82 | 7.13 | 217.19 | 6.48 | 0.02 |
| body_mass_g | 3700.66 | 458.57 | 3733.09 | 384.34 | 5076.02 | 504.12 | 0.26 |
Histograms
The datasummary family of functions allow users to
display in-line spark-style histograms to describe the distribution of
the variables. For example, the datasummary_skim produces
such a histogram:
tmp <- mtcars[, c("mpg", "hp")]
datasummary_skim(tmp)| Unique (#) | Missing (%) | Mean | SD | Min | Median | Max | ||
|---|---|---|---|---|---|---|---|---|
| mpg | 25 | 0 | 20.1 | 6.0 | 10.4 | 19.2 | 33.9 | |
| hp | 22 | 0 | 146.7 | 68.6 | 52.0 | 123.0 | 335.0 |
Each of the histograms in the table above is actually an SVG image,
produced by the kableExtra package. For this reason, the
histogram will not appear when users use a different output
backend, such as gt, flextable, or
huxtable.
The datasummary function is incredibly flexible, but it
does not include a histogram option by default. Here is a simple example
of how one can customize the output of datasummary. We
proceed in 4 steps:
- Normalize the variables and store them in a list
- Create the table with
datasummary, making sure to include 2 “empty” columns. In the example, we use a simple function calledemptycolto fill those columns with empty strings. - Add the histograms or boxplots using functions from the
kableExtrapackage.
tmp <- mtcars[, c("mpg", "hp")]
# create a list with individual variables
# remove missing and rescale
tmp_list <- lapply(tmp, na.omit)
tmp_list <- lapply(tmp_list, scale)
# create a table with `datasummary`
# add a histogram with column_spec and spec_hist
# add a boxplot with colun_spec and spec_box
emptycol = function(x) " "
datasummary(mpg + hp ~ Mean + SD + Heading("Boxplot") * emptycol + Heading("Histogram") * emptycol,
output = "kableExtra",
data = tmp) %>%
kableExtra::column_spec(column = 4, image = spec_boxplot(tmp_list)) %>%
kableExtra::column_spec(column = 5, image = spec_hist(tmp_list))If you want a simpler solution, you can try the
Histogram function which works in datasummary
automatically and comes bundled with modelsummary. The
downside of this function is that it uses Unicode characters to create
the histogram. This kind of histogram may not display well with certain
typefaces or on some operating systems (Windows!).
datasummary(mpg + hp ~ Mean + SD + Histogram, data = tmp)| Mean | SD | Histogram | |
|---|---|---|---|
| mpg | 20.09 | 6.03 | ▂▅▇▇▆▃▁▁▂▂ |
| hp | 146.69 | 68.56 | ▅▅▇▂▆▂▃▁▁ |
Missing values
At least 3 distinct issues can arise related to missing values.
Functions and na.rm
An empty cell can appear in the table when a statistical function
returns NA instead of a numeric value. In those cases, you
should:
- Make sure that your statistical function (e.g.,
meanorsd) usesna.rm=TRUEby default - Use the
Argumentsstrategy to setna.rm=TRUE(see theArgumentssection of this vignette). - Use a convenience function supplied by
modelsummary, wherena.rmisTRUEby default:Mean,SD,P25, etc.
Empty crosstab cells
An empty cell can appear in the table when a crosstab is deeply
nested, and there are no observations for a given combination of
covariates. In those cases, you can use the * DropEmpty
pseudo-function. See the “Empty cells” section of this vignette for
examples.
Factors
By default, the factor function in R does
not assign a distinct factor level to missing values: the
factor function’s exclude argument is set to
NA by default. To ensure that NAs appear in
your table, make sure you set exclude=NULL when you create
the factor.
Internally, the datasummary_balance and
datasummary_crosstab functions convert logical and
character variables to factor with the exclude=NULL
argument. This means that NAs will appear in the table as
distinct rows/columns. If you do not want NAs to
appear in your table, convert them to factors yourself ahead of time.
For example:
mycars <- mtcars[, c("cyl", "mpg", "hp", "vs")]
mycars$cyl[c(1, 3, 6, 8)] <- NA
mycars$cyl_nona <- factor(mycars$cyl)
mycars$cyl_na <- factor(mycars$cyl, exclude = NULL)
datasummary_crosstab(cyl_nona ~ vs, data = mycars)| cyl_nona | 0 | 1 | All | |
|---|---|---|---|---|
| 4 | N | 1 | 8 | 9 |
| % row | 11.1 | 88.9 | 100.0 | |
| 6 | N | 2 | 3 | 5 |
| % row | 40.0 | 60.0 | 100.0 | |
| 8 | N | 14 | 0 | 14 |
| % row | 100.0 | 0.0 | 100.0 | |
| All | N | 18 | 14 | 32 |
| % row | 56.2 | 43.8 | 100.0 |
datasummary_crosstab(cyl_na ~ vs, data = mycars)| cyl_na | 0 | 1 | All | |
|---|---|---|---|---|
| 4 | N | 1 | 8 | 9 |
| % row | 11.1 | 88.9 | 100.0 | |
| 6 | N | 2 | 3 | 5 |
| % row | 40.0 | 60.0 | 100.0 | |
| 8 | N | 14 | 0 | 14 |
| % row | 100.0 | 0.0 | 100.0 | |
| NA | N | 1 | 3 | 4 |
| % row | 25.0 | 75.0 | 100.0 | |
| All | N | 18 | 14 | 32 |
| % row | 56.2 | 43.8 | 100.0 |
Appearance
In the Appearance
Vignette we saw how to customize the tables produced by the
modelsummary function. The same customization possibilities
are also available for all functions in the datasummary_*
family of functions. Indeed, we can customize the tables produced by
datasummary using the functions provided by
gt, kableExtra, flextable,
huxtable, and DT. For instance, to customize
tables using kableExtra, we can do:
datasummary(All(penguins) ~ sex * (Mean + SD),
data = penguins,
output = 'kableExtra') %>%
kableExtra::row_spec(3, background = 'cyan', color = 'red')To customize a table using the gt package, we can:
library(gt)
adelie <- function(x) web_image('https://user-images.githubusercontent.com/987057/85402702-20b1d280-b52a-11ea-9950-f3a03133fd45.png', height = 100)
gentoo <- function(x) web_image('https://user-images.githubusercontent.com/987057/85402718-27404a00-b52a-11ea-9ad3-dd7562f6438d.png', height = 100)
chinstrap <- function(x) web_image('https://user-images.githubusercontent.com/987057/85402708-23acc300-b52a-11ea-9a77-de360a0d1f7d.png', height = 100)
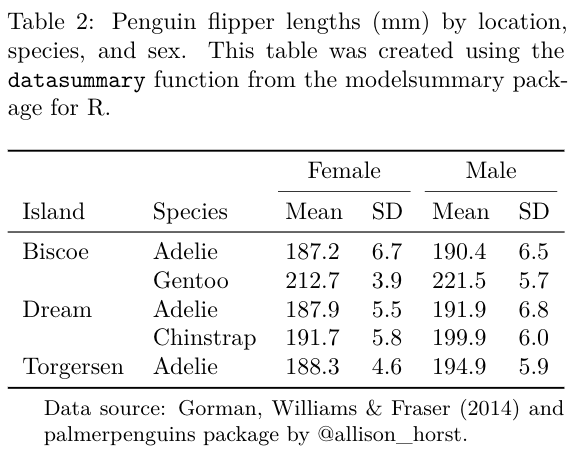
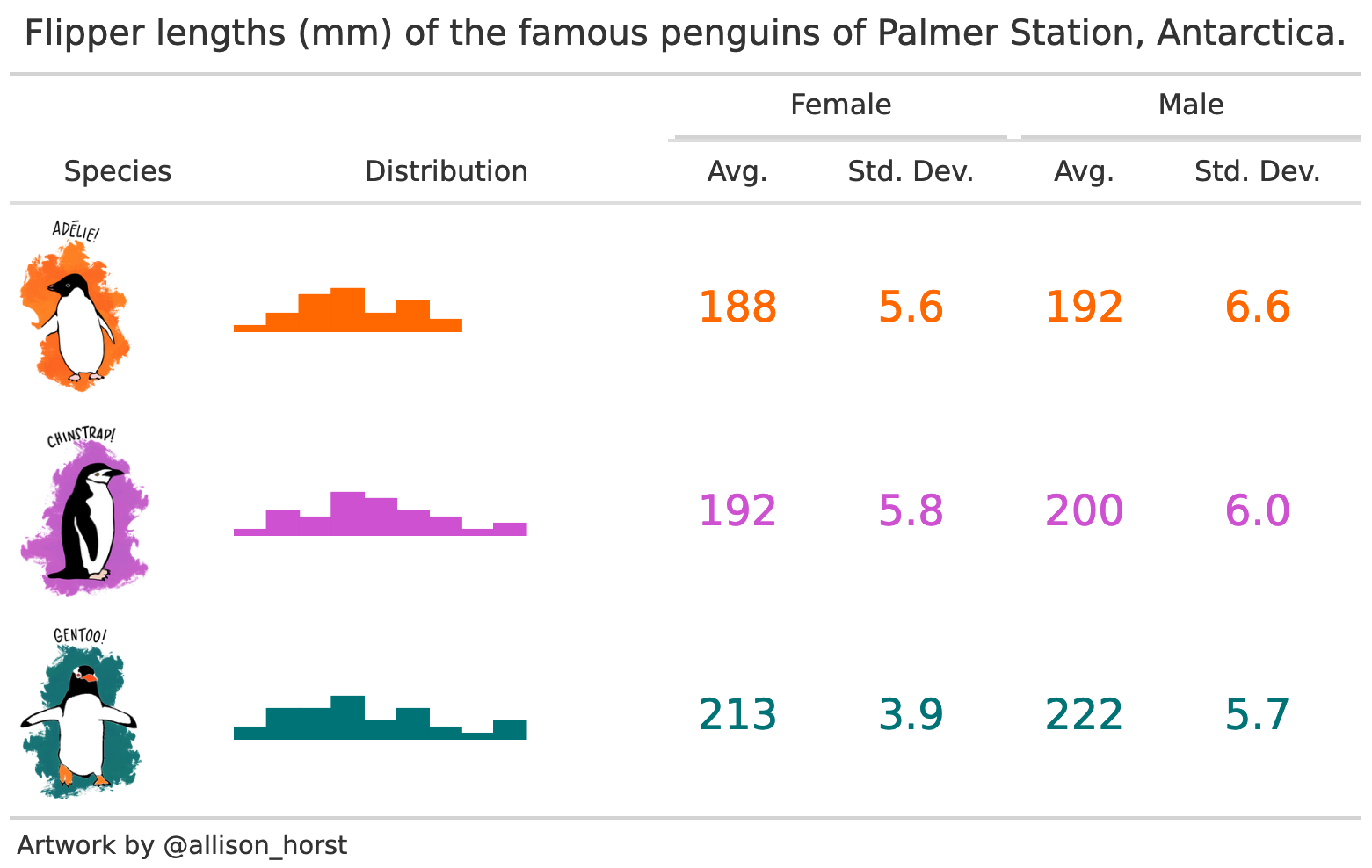
cap <- 'Flipper lengths (mm) of the famous penguins of Palmer Station, Antarctica.'
f <- (`Species` = species) ~ (` ` = flipper_length_mm) * (`Distribution` = Histogram) + flipper_length_mm * sex * ((`Avg.` = Mean)*Arguments(fmt='%.0f') + (`Std. Dev.` = SD)*Arguments(fmt='%.1f'))
datasummary(f,
data = penguins,
output = 'gt',
title = cap,
notes = 'Artwork by @allison_horst',
sparse_header = TRUE) %>%
text_transform(locations = cells_body(columns = 1, rows = 1), fn = adelie) %>%
text_transform(locations = cells_body(columns = 1, rows = 2), fn = chinstrap) %>%
text_transform(locations = cells_body(columns = 1, rows = 3), fn = gentoo) %>%
tab_style(style = list(cell_text(color = "#FF6700", size = 'x-large')), locations = cells_body(rows = 1)) %>%
tab_style(style = list(cell_text(color = "#CD51D1", size = 'x-large')), locations = cells_body(rows = 2)) %>%
tab_style(style = list(cell_text(color = "#007377", size = 'x-large')), locations = cells_body(rows = 3)) %>%
tab_options(table_body.hlines.width = 0, table.border.top.width = 0, table.border.bottom.width = 0) %>%
cols_align('center', columns = 3:6)| Species | Distribution | Female | Male | ||
|---|---|---|---|---|---|
| Avg. | Std. Dev. | Avg. | Std. Dev. | ||
 |
▁▃▆▇▃▅▂ | 188 | 5.6 | 192 | 6.6 |
 |
▁▄▃▇▆▄▃▁▂ | 192 | 5.8 | 200 | 6.0 |
 |
▂▅▅▇▃▅▂▁▃ | 213 | 3.9 | 222 | 5.7 |
| Artwork by @allison_horst | |||||